Running experiments inside a Jupyter Notebook
- Aug. 29, 2023
Researchers on Chameleon are working on interesting problems in computer science. For research to be impactful, it should be reproducible by others in the field, and easily extended when new researchers are inspired by the original results. This is why on Chameleon we are working to provide tools for practical reproducibility. There are three parts of practical reproducibility are packaging, sharing, and access (you can read more in our paper). Sharing and access are addressed on Chameleon with Trovi and Daypass respectively. The packaging of an experiment is the form in which the configuration, program, and benchmarks run, along with the data and tools for its analysis. We’ve written before in more detail about how to do each of these steps on Chameleon’s JupyterHub. If you would like to see examples of how different types of experiments look packaged for this, you see existing experiment patterns on Trovi.
These examples typically set up and configure resources via python-chi, and once their instance(s) are running, they run commands on it via ssh to execute the experiment and collect data all from within a notebook running on Chameleon’s JupyterHub. This data is copied off of the instance, and then the notebook can analyze it, typically outputting some graph to share eventually in a paper. There are 2 limitations of this workflow:
- Rather than run via SSH, some researchers write their experimental program as a notebook, and want to run it directly on the instance to make use of its CPU, RAM, or GPU.
- Chameleon’s JupyterHub runs notebooks in a low power environment, with only 2 CPU threads, a few GB of RAM, and only a GB of storage. Depending on your data, this may not be a sufficient environment in which to analyze it.
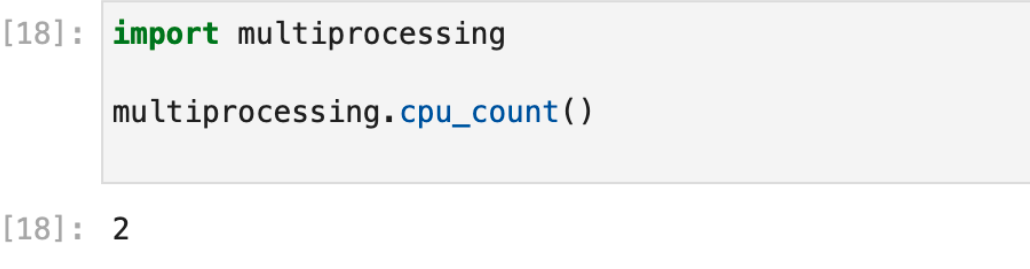
The limited multiprocessing capabilities of Chameleons’ JupyterHub
To get around these limitations, we’ve created the Basic Jupyter Server artifact, which demonstrates how to install and connect to a secure Jupyter environment on a Chameleon instance. The interesting parts of this process, which should work on any Chameleon instance, are:
- Install the jupyter-notebook package on an instance, and configure it.
- Start a service to run the notebook server with.
- Connect to your instance via an SSH tunnel from your laptop or desktop computer.
Once this is done, you should see the Jupyter notebook home screen. You can select “new > terminal” and execute “lscpu” (or “nvidia-smi”) to confirm that it is running on a powerful Chameleon node.
You can augment the installation of jupyter-notebook to copy your experiment or data analysis notebooks to the node. Then upon opening them, you’ll see the notebook kernel is running directly on the instance, as expected. If your instance has a GPU, this will enable you to use it with CUDA inside the notebook.
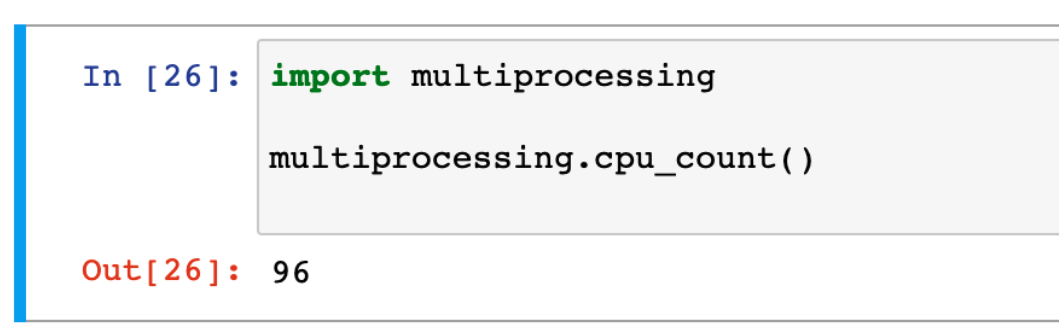
Powerful multiprocessing capabilities of Jupyter running on a Chameleon node
Using this method, your Jupyter notebooks have the full power of Chameleon’s hardware. You could now access low level properties about the machine, train a machine learning model, or stitch to Fabric all natively in your notebooks python kernel. If you are feeling adventurous, you could even take a Colab notebook, and upload it to this open environment (some translation may be required, your instance won’t have the same pre-installed libraries). The possibilities are endless!
No comments